DigitalBiology 1.01: Introduction to Bioinformatics
Welcome to the world of Digital Biology! This guide is designed to introduce you to the fascinating intersection of biology and computer science. Whether you are a biologist learning to code or a developer diving into life sciences, this overview will set the stage for your journey.
1. Introduction: From Bioinformatics to Digital Biology
### What is Bioinformatics?
At its core, Bioinformatics is the science of collecting, processing, storing, analyzing, and interpreting complex biological data. It acts as a bridge between Information Science (Computer Science) and Biology.
- The Goal: To reveal the secrets hidden within massive and complex biological datasets (like the human genome) using computational methods.
- The Structure: It is an interdisciplinary field combining molecular biology, genetics, computer science, mathematics, and statistics.
- The Distinction: You might hear "Computational Biology" used interchangeably. Strictly speaking, Bioinformatics focuses on the methods and tools to handle data (Biology + Informatics), while Computational Biology focuses on using these tools to study biological questions. However, in practice, the lines are blurred.
### The Era of "Digital Biology"
We are entering a new era often described as Digital Biology. This concept goes beyond just "analyzing data"; it treats biology as an information system that can be modeled, simulated, and even programmed.
Jensen Huang (CEO of NVIDIA) has been a vocal proponent of this shift. He describes digital biology as potentially "one of the biggest technology revolutions ever."

"For the very first time in our history, biology has the opportunity to become engineering, not science. When something becomes engineering, it becomes less sporadic and exponentially improving." — Jensen Huang
In this view, the "code of life" (DNA) is literally code. With the rise of AI and Generative models (like AlphaFold), we are moving from just reading biology to understanding and designing it. AI is accelerating this field exponentially, helping us design new proteins, drugs, and therapies in silico (on a computer) before ever touching a test tube.
2. A History of Revolution (Timeline)
The history of bioinformatics is deeply tied to the history of sequencing technology — our ability to "read" DNA.
### Early Foundations (Pre-2000)
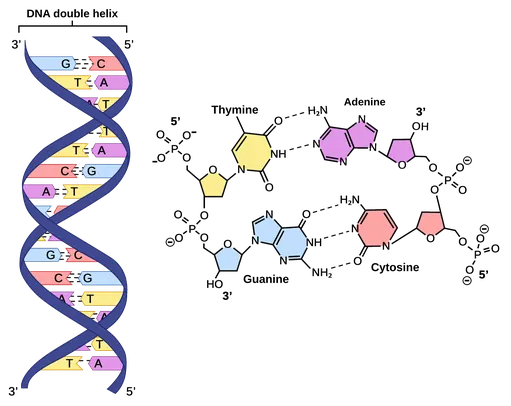
- 1953: Discovery of the DNA Double Helix (Watson & Crick).
- 1977: Sanger Sequencing invented. The first method to read DNA, but it was slow and expensive.
- 1990–2003: The Human Genome Project. A massive 13-year international effort to sequence one human genome for nearly $3 billion.
### The NGS Explosion (2005–2015)
This era saw the rise of Next-Generation Sequencing (NGS), making data generation massive, cheap, and fast.
Year 2005:
- HapMap Project completed (mapping human genetic variation).
- 454 Life Sciences releases the first "Next-Gen" sequencer.
- Oxford Nanopore is founded (pioneering portable sequencing).
Year 2006:
- Solexa (later acquired by Illumina) releases the Genome Analyzer, capable of sequencing 1 Gb of data in a single run. This was a game-changer.
Year 2007:
- First "Personal Genome" completed.
- Roche acquires 454; ABI launches SOLiD sequencer.
Year 2008:
- 1000 Genomes Project launches to map genetic diversity across humanity.
Year 2010:
- BGI Metagenomics Breakthrough: BGI published a landmark paper in Nature ("A human gut microbial gene catalogue established by metagenomic sequencing"), proving that NGS could be used to sequence complex microbial communities (metagenomics) at scale. This shifted the field from studying single organisms to entire ecosystems.
- Illumina HiSeq 2000 released. The dominance of Illumina in the market begins.
- NIPT (Non-Invasive Prenatal Testing) enters the clinic.
Between 2011 and 2012:
- Oxford Nanopore reveals the first data from a "nanopore" sequencer.
- Illumina HiSeq 2500 and MiSeq released.
Year 2014:
- Illumina HiSeq X Ten. The factory-scale sequencer that promised the "$1000 Genome," making population-scale sequencing a reality. "MinION" early access program begins for portable sequencing.
### The Modern Era: Precision & Scale (2015 — Present)
Between 2015 and 2020:
- Precision Medicine Initiative announced. Liquid Biopsy becomes a major focus.
- 10x Genomics launches the Chromium platform, kicking off the Single-Cell Sequencing revolution.
- Illumina NovaSeq released, driving costs down further. BGI (MGI) releases competitive sequencers (MGISEQ-2000).
2020 — Present:
- Nanopore Sequencing hits high accuracy (98.3%+).
- AI & Large Language Models begin to revolutionize protein folding (AlphaFold) and genomic interpretation.
2024:
- BGI releases CycloneSEQ nanopore sequencer, further diversifying the market.
3. Major Applications
Bioinformatics is the engine behind modern biological discoveries. It is applied across three main fields:
### A. Medical Research (Precision Medicine)
- NIPT (Non-Invasive Prenatal Testing): Screening fetal health from maternal blood.
- Tumor Genomics: Identifying specific mutations in cancer to guide targeted therapy (e.g., EGFR inhibitors).
- PGS (Pre-implantation Genetic Screening): Selecting healthy embryos during IVF.
- Rare & Complex Disease Diagnosis: Finding the "needle in the haystack" mutation for undiagnosed patients.
- DTC (Direct-to-Consumer): Tests like 23andMe for ancestry and health risks.
- Pathogen Detection: Rapidly identifying viruses (like SARS-CoV-2) or bacteria in clinical samples.
### B. Agricultural Research
- Molecular Breeding: Using genetic markers to select for better crops (higher yield, drought resistance) without waiting for them to grow.
- Trait Mapping (GWAS): Finding which genes control specific traits (e.g., fruit size, milk production).
- Population Evolution: Understanding the genetic diversity and history of livestock and crops.
### C. Basic Scientific Research
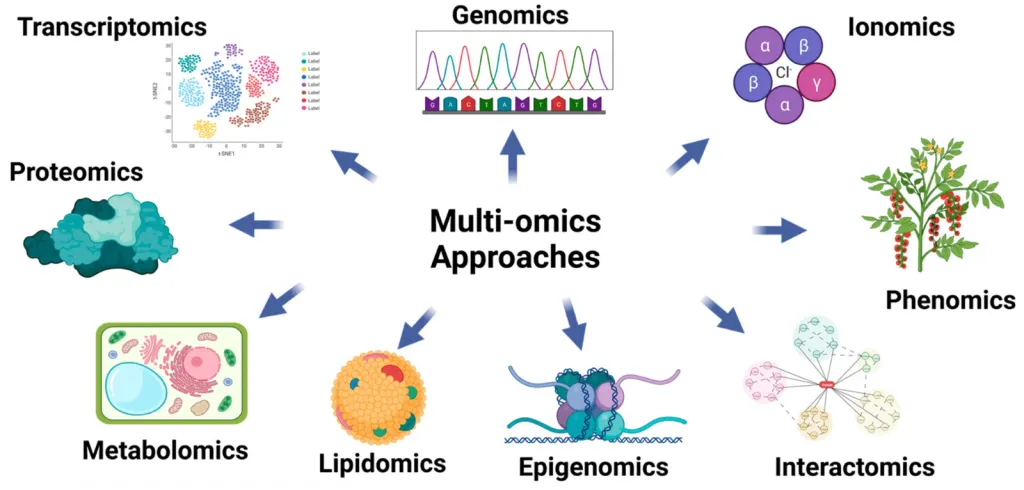
- Multi-Omics: Integrating data from Genomics (DNA), Transcriptomics (RNA), Proteomics (Protein), and Epigenomics (Methylation) to get a complete picture of life.
- Metagenomics: Studying microbial communities in the gut, soil, or ocean.
- Single-Cell Sequencing: Analyzing biology one cell at a time to understand heterogeneity.
4. Why is it Hard? (Challenges)
Bioinformatics is notoriously difficult for beginners. Based on student experiences, here are the three main reasons:
- The "Snowball" of Interdisciplinary Skills: It is not just learning one thing. You need Biology (to understand the problem) + Computer Science (to run the tools) + Statistics (to interpret the results). Many students fail because they try to learn "just the coding" without the stats, or "just the biology" without the Linux skills. It requires a "hybrid" brain.
- Breadth of Content: Biology is vast. Analyzing DNA (static code) is completely different from analyzing RNA (dynamic levels) or Proteins (3D structures). Techniques differ wildly between species (Plants vs. Humans vs. Bacteria). There is no "one size fits all" workflow.
- Rapid Technological Evolution: The technology changes faster than textbooks. We moved from Sanger -> Illumina (Short reads) -> PacBio/Nanopore (Long reads). Each platform produces different data with different error profiles. A tool you learned 3 years ago might be obsolete today. You must be a lifelong learner.
5. Common Misunderstandings & Corrections
Let's address the top questions and myths from beginners:
- "Can I publish papers just by knowing bioinformatics?" — Reality: Yes, but it's getting harder. Pure data mining papers exist, but high-impact work usually requires experimental validation.
- "Do I really need to know the sequencing principle?" — Reality: YES. If you don't understand how the data was generated (e.g., PCR bias, sequencing errors), you will misinterpret the results (artifacts vs. real biology).
- "Is bioinformatics just running a pipeline?" — Reality: No. Running the pipeline is the easy part (10%). Interpreting the result, troubleshooting failed steps, and customizing parameters for your specific data is the hard part (90%).
- "Is the core of bioinformatics learning algorithms?" — Reality: For developers, yes. For analysts (most of us), the core is biological interpretation and data logic, not writing new alignment algorithms from scratch.
- "Will 3rd Generation Sequencing (Long Read) replace 2nd Gen (Illumina)?" — Reality: Not immediately. They have different strengths. Illumina is still cheaper and more accurate for simple counting (RNA-Seq). They will coexist for a long time.
- "Is doing bioinformatics 'better' than wet lab?" — Reality: It's not better, just different. Dry lab avoids toxic chemicals but introduces "toxic" data bugs. The best scientists often understand both.